The file babies.csv contains data on baby birthweights. The variables are:
babyData = read.csv('babies.csv')
head(babyData)
dim(babyData)| case | bwt | gestation | parity | age | height | weight | smoke | |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 120 | 284 | 0 | 27 | 62 | 100 | 0 |
| 2 | 2 | 113 | 282 | 0 | 33 | 64 | 135 | 0 |
| 3 | 3 | 128 | 279 | 0 | 28 | 64 | 115 | 1 |
| 4 | 4 | 123 | NA | 0 | 36 | 69 | 190 | 0 |
| 5 | 5 | 108 | 282 | 0 | 23 | 67 | 125 | 1 |
| 6 | 6 | 136 | 286 | 0 | 25 | 62 | 93 | 0 |
There are a lot of cells with NA (not available) entries, and these could mess up our analysis below. The na.omit() command is a fast way to remove these.
babyData = na.omit(babyData)
head(babyData)
dim(babyData)| case | bwt | gestation | parity | age | height | weight | smoke | |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 120 | 284 | 0 | 27 | 62 | 100 | 0 |
| 2 | 2 | 113 | 282 | 0 | 33 | 64 | 135 | 0 |
| 3 | 3 | 128 | 279 | 0 | 28 | 64 | 115 | 1 |
| 5 | 5 | 108 | 282 | 0 | 23 | 67 | 125 | 1 |
| 6 | 6 | 136 | 286 | 0 | 25 | 62 | 93 | 0 |
| 7 | 7 | 138 | 244 | 0 | 33 | 62 | 178 | 0 |
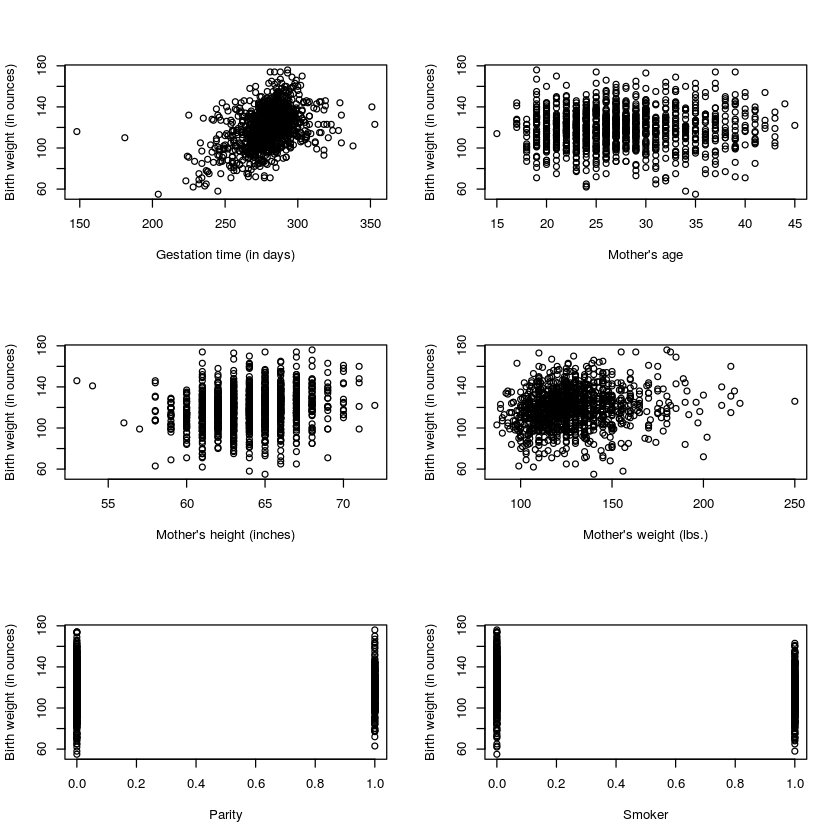
We need to check that there is a roughly linear relationship between each of the explanatory variables and the response variable. The par() command below lets us arrange the graphs in a 3-by-2 matrix.
par(mfrow=c(3,2))
plot(babyData$gestation,babyData$bwt,xlab='Gestation time (in days)',ylab='Birth weight (in ounces)')
plot(babyData$age,babyData$bwt,xlab="Mother's age",ylab='Birth weight (in ounces)')
plot(babyData$height,babyData$bwt,xlab="Mother's height (inches)",ylab='Birth weight (in ounces)')
plot(babyData$weight,babyData$bwt,xlab="Mother's weight (lbs.)",ylab='Birth weight (in ounces)')
plot(babyData$parity,babyData$bwt,xlab="Parity",ylab='Birth weight (in ounces)')
plot(babyData$smoke,babyData$bwt,xlab="Smoker",ylab='Birth weight (in ounces)')
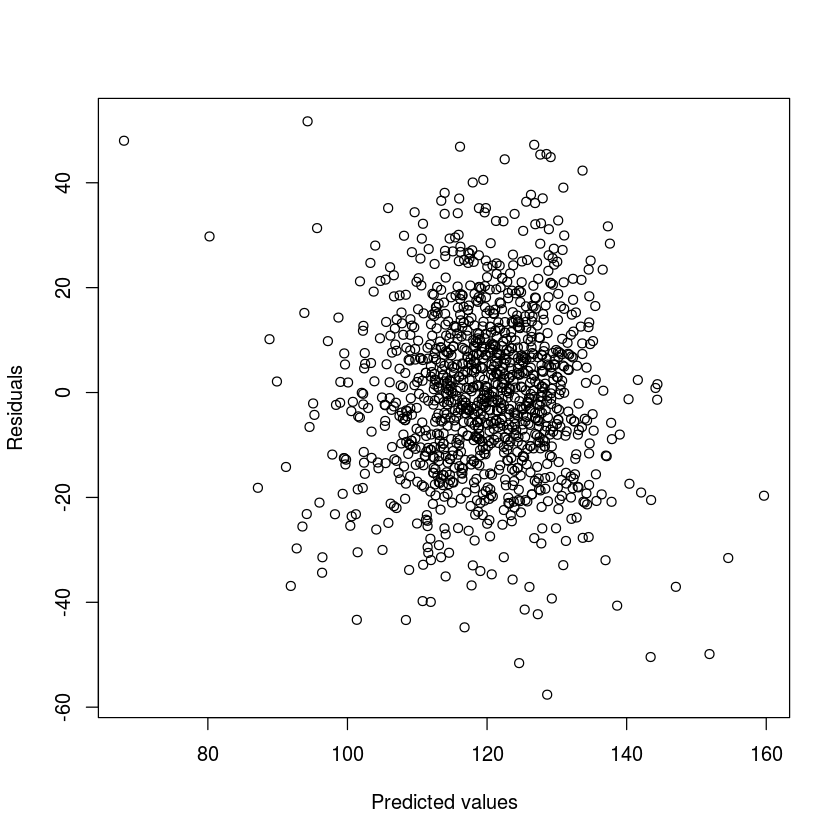
Just like in single variable regression, we need to check the residuals to see that they are roughly normally distributed with the same variance. This is much harder to do with so many variables. So here are some of the most important cases to check:
myLM = lm(bwt~gestation+age+height+weight+parity+smoke,data=babyData)qqnorm(resid(myLM))
plot(fitted(myLM),resid(myLM),xlab='Predicted values',ylab='Residuals')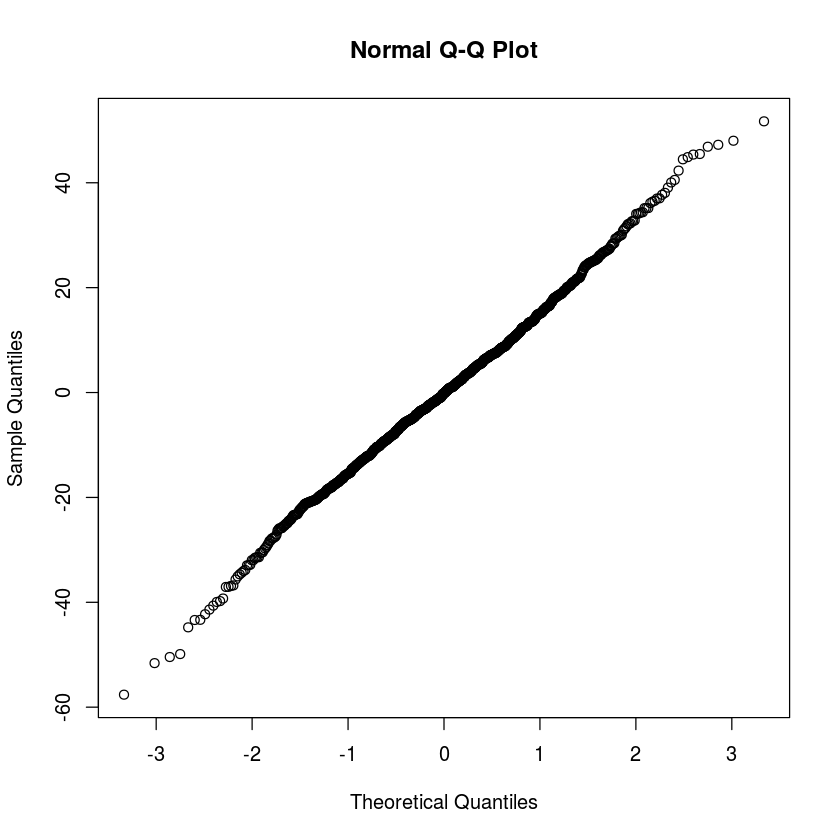
par(mfrow=c(3,2))
plot(babyData$gestation,resid(myLM),xlab='Gestation time (in days)',ylab='Residuals')
plot(babyData$age,resid(myLM),xlab="Mother's age",ylab='Residuals')
plot(babyData$height,resid(myLM),xlab="Mother's height (inches)",ylab='Residuals')
plot(babyData$weight,resid(myLM),xlab="Mother's weight (lbs.)",ylab='Residuals')
plot(babyData$parity,resid(myLM),xlab="Parity",ylab='Residuals')
plot(babyData$smoke,resid(myLM),xlab="Smoker",ylab='Residuals')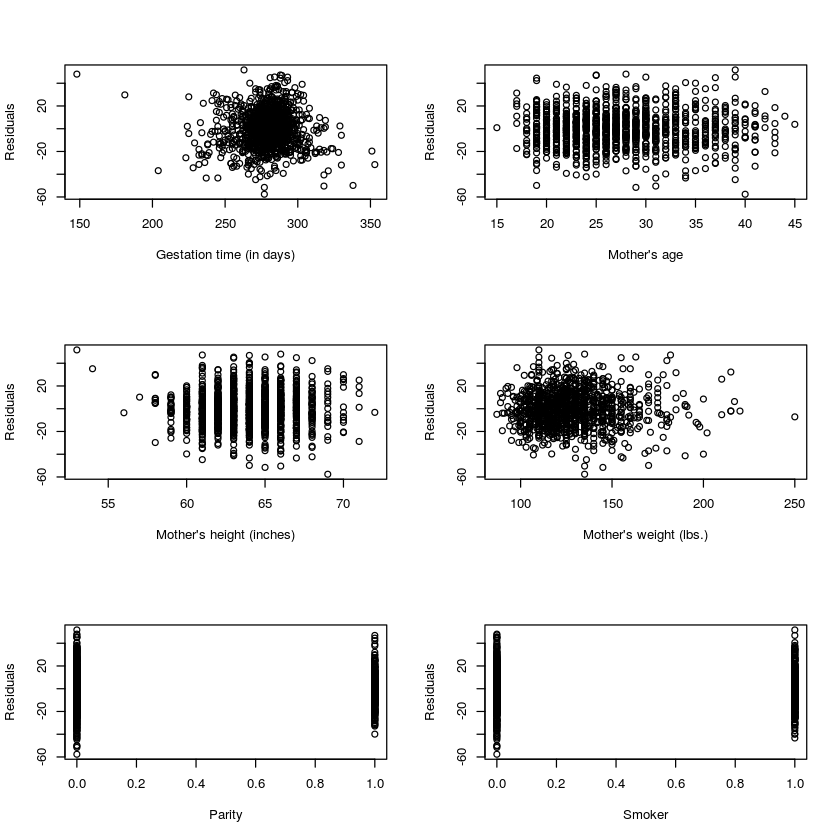
The residuals mostly seem to have the same variance throughout, there is not clear trend in the scatterplots above. The qq-plot make it clear that the residuals are very normal, which is good.
summary(myLM)Call:
lm(formula = bwt ~ gestation + age + height + weight + parity +
smoke, data = babyData)
Residuals:
Min 1Q Median 3Q Max
-57.613 -10.189 -0.135 9.683 51.713
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -80.41085 14.34657 -5.605 2.60e-08 ***
gestation 0.44398 0.02910 15.258 < 2e-16 ***
age -0.00895 0.08582 -0.104 0.91696
height 1.15402 0.20502 5.629 2.27e-08 ***
weight 0.05017 0.02524 1.987 0.04711 *
parity -3.32720 1.12895 -2.947 0.00327 **
smoke -8.40073 0.95382 -8.807 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 15.83 on 1167 degrees of freedom
Multiple R-squared: 0.258, Adjusted R-squared: 0.2541
F-statistic: 67.61 on 6 and 1167 DF, p-value: < 2.2e-16We will now remove variables from the full model to get the model with the best adjusted R-squared.
adjustedLM = lm(bwt~gestation+height+weight+parity+smoke,data=babyData)
summary(adjustedLM)Call:
lm(formula = bwt ~ gestation + height + weight + parity + smoke,
data = babyData)
Residuals:
Min 1Q Median 3Q Max
-57.716 -10.150 -0.159 9.689 51.620
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -80.71321 14.04465 -5.747 1.16e-08 ***
gestation 0.44408 0.02907 15.276 < 2e-16 ***
height 1.15497 0.20473 5.641 2.11e-08 ***
weight 0.04983 0.02503 1.991 0.04672 *
parity -3.28762 1.06281 -3.093 0.00203 **
smoke -8.39390 0.95117 -8.825 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 15.82 on 1168 degrees of freedom
Multiple R-squared: 0.2579, Adjusted R-squared: 0.2548
F-statistic: 81.2 on 5 and 1168 DF, p-value: < 2.2e-16These work exactly the same as the single variable case.
confint(adjustedLM)| 2.5 % | 97.5 % | |
|---|---|---|
| (Intercept) | -108.26876 | -53.15765 |
| gestation | 0.3870403 | 0.5011121 |
| height | 0.753293 | 1.556649 |
| weight | 0.000724759 | 0.098942229 |
| parity | -5.372856 | -1.202391 |
| smoke | -10.260090 | -6.527717 |
predict(adjustedLM,data.frame(gestation = 240,height=70,weight=120,age=25,parity=1,smoke=0),interval='prediction')| fit | lwr | upr | |
|---|---|---|---|
| 1 | 109.40544 | 78.10146 | 140.70942 |